Molecule Discovery: A Key to Scientific Advancement
Understanding the Challenges
Molecule discovery is crucial in fields like pharmaceuticals and materials science. While Graph Neural Networks (GNNs) have improved how we represent molecules and predict their properties, they struggle to adapt across different tasks and often require a lot of specific data. Additionally, generating molecules with specific properties remains a challenge. Integrating Large Language Models (LLMs) into this process also faces obstacles, such as aligning molecular and textual data and dealing with limited datasets.
AI Solutions for Molecule Discovery
To enhance molecule discovery, various AI techniques have been developed. These include:
– **Machine Learning and Deep Learning**: These methods allow for complex analysis of biological and chemical data.
– **Convolutional Neural Networks (CNNs)**: Useful for analyzing molecular structures.
– **Recurrent Neural Networks (RNNs)**: Effective for processing sequential data.
– **Transformer-based Networks**: Excellent for recognizing complex patterns.
A notable approach is **Text-based Molecule Generation (Text2Mol)**, which uses natural language descriptions to retrieve molecules. Models like **MolT5** have shown promise in generating SMILES strings, while advancements like **KVPLM**, **MoMu**, and **3DMoLM** have improved capabilities by utilizing molecular graphs and spatial configurations.
Introducing TOMG-Bench
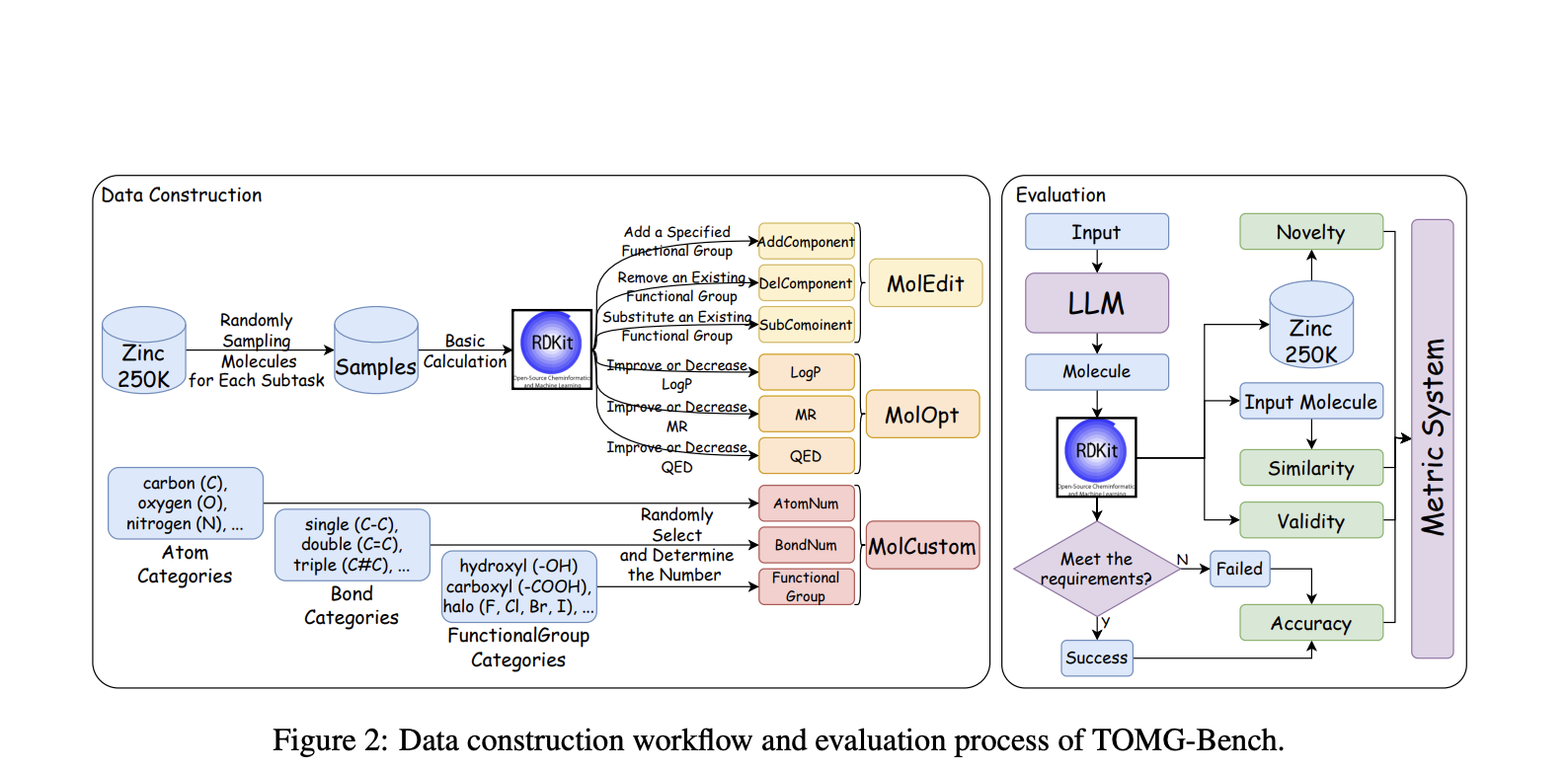
Researchers from The Hong Kong Polytechnic University, Shanghai Jiao Tong University, and Shanghai AI Lab have created **TOMG-Bench**, the first comprehensive benchmark for evaluating LLMs in open-domain molecule generation. It includes three main tasks:
– **Molecule Editing (MolEdit)**
– **Molecule Optimization (MolOpt)**
– **Customized Molecule Generation (MolCustom)**
Each task has three subtasks with 5,000 test samples. An automated evaluation system assesses the quality and accuracy of the generated molecules, providing valuable insights into the limitations of current text-guided molecule discovery.
Evaluation Framework
TOMG-Bench evaluates four categories of models:
1. **Proprietary Models**: Commercial systems like GPT-4-turbo and Claude-3.5.
2. **Open-source General LLMs**: Models like Llama-3 and Mistral-7B.
3. **Fine-tuned LLMs on ChEBI-20**: Including MolT5 and BioT5-base.
4. **OpenMolIns Fine-tuned LLMs**: Featuring Galactica-125M and others.
The evaluation revealed that **Claude-3.5** performed best with an accuracy of 35.92%, followed by **Gemini-1.5-pro** at 34.80%. Open-source models like **Llama-3-70B-Instruct** showed significant progress, achieving 23.93% accuracy.
Implications of TOMG-Bench
TOMG-Bench highlights both the limitations and potential of LLMs in molecule generation. While some models show promise, challenges remain, such as insufficient diversity in prompts and inaccuracies in molecular component distributions.
Get Involved and Evolve with AI
Explore how AI can transform your business. Here are some practical steps:
– **Identify Automation Opportunities**: Find areas in customer interactions that can benefit from AI.
– **Define KPIs**: Ensure your AI initiatives have measurable impacts.
– **Select an AI Solution**: Choose tools that fit your needs and allow customization.
– **Implement Gradually**: Start small, gather data, and expand wisely.
For AI KPI management advice, connect with us at hello@itinai.com. For ongoing insights, follow us on Telegram or @itinaicom.
Discover how AI can enhance your sales processes and customer engagement at itinai.com.


























