Challenges in Modeling Biological and Chemical Sequences
Modeling biological and chemical sequences is complex due to long-range dependencies and the need to process large data efficiently. Traditional methods, especially Transformer-based architectures, struggle with long genomic sequences and protein modeling because they are slow and expensive to compute. Additionally, these models often cannot adapt to new tasks without retraining, which limits their effectiveness in fields like genomics, protein engineering, and drug discovery.
Current Limitations
Most existing methods rely on Transformer architectures, which are powerful but costly due to their self-attention mechanisms. While state-space models like S4 and Mamba offer some improvements, they still face issues with flexibility and high computational costs. Transformers also have short context windows, making them less efficient for long sequences, such as DNA analysis and protein folding. These challenges can hinder real-time applications and scalability.
Introducing Bio-xLSTM
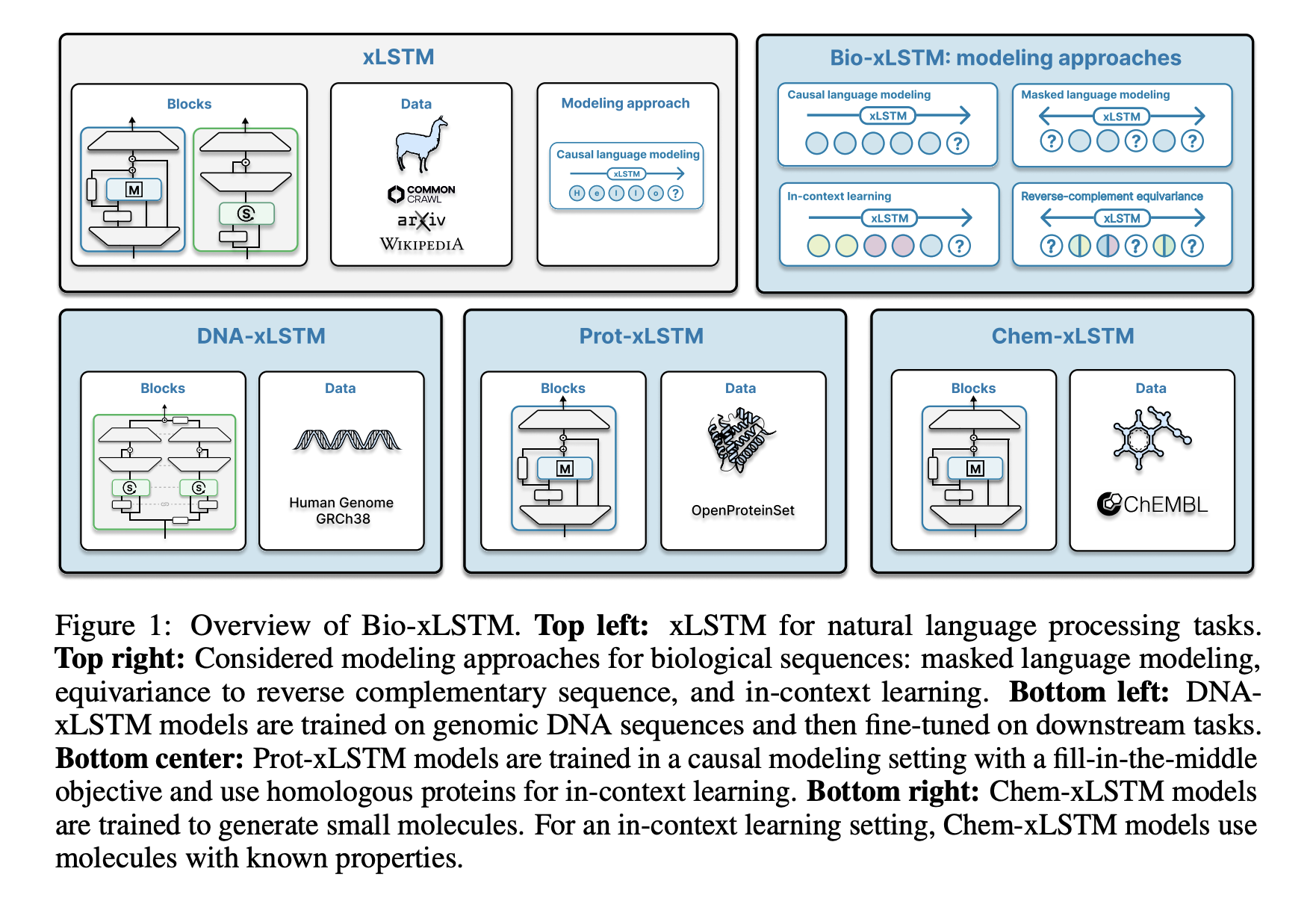
To address these issues, researchers from Johannes Kepler University and NXAI GmbH have developed Bio-xLSTM, an xLSTM variant designed specifically for biological and chemical sequences. Unlike Transformers, Bio-xLSTM processes sequences with linear runtime complexity, making it significantly more efficient. Key innovations include:
- DNA-xLSTM: Optimized for genomic sequences using reverse-complement equivariant mechanisms.
- Prot-xLSTM: Focused on protein prediction, utilizing homologous protein information.
- Chem-xLSTM: Designed for small molecule synthesis, supporting in-context learning.
Benefits of Bio-xLSTM
Bio-xLSTM utilizes specialized mechanisms and improved memory components for efficient sequence processing. It can handle large-scale genomic, protein, and chemical databases, supporting effective pre-training and fine-tuning. Each variant is tailored for its domain while maintaining the efficiency of xLSTM.
Performance Advantages
Bio-xLSTM significantly outperforms existing models in various tasks:
- In DNA sequence tasks, it shows lower validation loss compared to Transformer-based and state-space models.
- For protein modeling, it excels in generating homology-aware sequences with lower perplexity.
- In chemical sequences, it produces chemically valid structures with high accuracy.
A Game-Changer in Sequence Modeling
Bio-xLSTM is a revolutionary solution for biological and chemical sequence modeling. Its ability to overcome the limitations of Transformers and its domain-specific adaptations offer a scalable and effective approach for DNA, protein, and small molecule modeling. This positions Bio-xLSTM as a foundational tool in molecular biology and drug discovery, paving the way for more efficient AI-driven research in life sciences.
Get Involved
Check out the Paper for more details on this research. Follow us on Twitter, join our Telegram Channel, and be a part of our LinkedIn Group. Don’t forget to join our 75k+ ML SubReddit.
Embrace AI for Your Business
Transform your company with AI using Bio-xLSTM. Here’s how:
- Identify Automation Opportunities: Find key customer interaction points that can benefit from AI.
- Define KPIs: Ensure measurable impacts on business outcomes.
- Select an AI Solution: Choose tools that match your needs and allow customization.
- Implement Gradually: Start with a pilot program, gather data, and expand wisely.
For AI KPI management advice, connect with us at hello@itinai.com. Stay updated on leveraging AI through our Telegram at t.me/itinainews or Twitter @itinaicom.
Discover how AI can enhance your sales processes and customer engagement. Explore solutions at itinai.com.



























