Machine Learning Models for Predicting Prime Editing Efficiency
Practical Solutions and Value
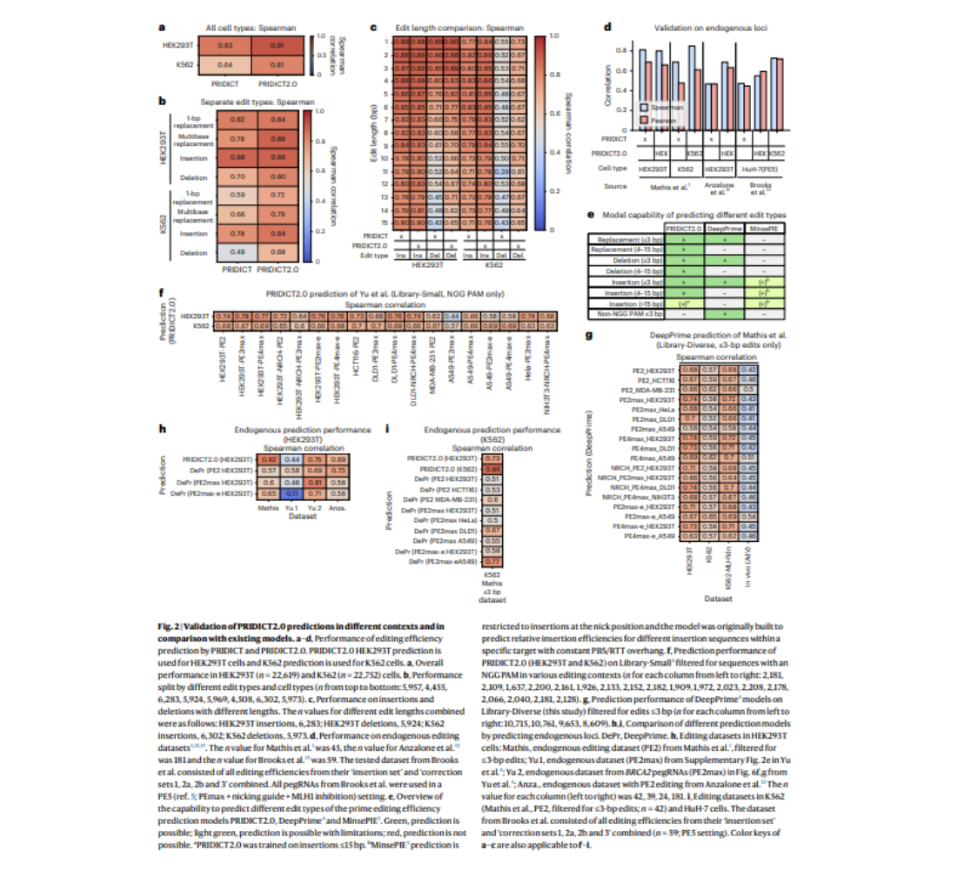
The success of prime editing relies on pegRNA design and target locus. PRIDICT2.0 and ePRIDICT are machine learning models that predict prime editing efficiency across various edit types and chromatin contexts. PRIDICT2.0 assesses pegRNA performance for edits up to 15 base pairs in MMR-deficient and proficient cell lines, while ePRIDICT quantifies how local chromatin environments impact prime editing rates. These models offer valuable tools for improving pegRNA design and maximizing the efficiency of prime editing in diverse biological contexts.
Insights into Chromatin Context and Editing Efficiency
Practical Solutions and Value
ePRIDICT predicts editing outcomes by accounting for locus-specific chromatin features, adding a new layer of precision to editing predictions. The models offer valuable tools for improving pegRNA design and maximizing the efficiency of prime editing in diverse biological contexts.
Chromatin’s Role in Genome Editing Efficiency
Practical Solutions and Value
Chromatin features influence genome editing efficiency. The ‘ePRIDICT’ model, trained on chromatin data, effectively predicts editing outcomes. Combining it with PRIDICT2.0 improves accuracy, especially in regions with lower chromatin accessibility, confirming chromatin’s pivotal role in editing outcomes.
Cloning and PegRNA Design
Practical Solutions and Value
PegRNAs were designed to achieve various genetic modifications and cloned using a particular vector. This process ensures the efficient design and implementation of pegRNAs for prime editing.
Viral Vector Production and Screening
Practical Solutions and Value
Lentiviral and pseudotyped AAV9 vectors were produced and used for editing in various cell lines. The screening process involved transducing cells with lentivirus and selecting edited cells using antibiotics. This method provides a practical approach for editing efficiency analysis.
Library and Editing Efficiency Analysis
Practical Solutions and Value
Sequencing reads were analyzed with custom scripts and cross-referenced with chromatin data from ENCODE. Machine learning models, including PRIDICT2.0, were trained and validated using various datasets, providing insights into editing efficiency and guiding future experiments.



























