Introduction to Protein Design and Deep Learning
Protein design and prediction are essential for advancements in synthetic biology and therapeutics. While deep learning models like AlphaFold and ProteinMPNN have made great strides, there is a lack of accessible educational resources. This gap limits the understanding and application of these technologies.
The challenge is to create practical tools that help researchers, educators, and students apply deep learning techniques in protein design, combining theoretical knowledge with real-world applications.
DL4Proteins Notebook Series
DL4Proteins is a series of Jupyter notebooks developed by Graylab researchers to make deep learning in protein design more accessible. Inspired by the 2024 Nobel Prize winners, this series offers practical introductions to tools like AlphaFold and ProteinMPNN.
Target Audience: Researchers, educators, and students will benefit from this resource, which combines foundational machine learning concepts with advanced protein engineering methods.
Notebooks Overview
- Neural Networks with NumPy: Understand basic neural network concepts using NumPy. This notebook emphasizes core operations like matrix multiplication, making it ideal for beginners.
- Neural Networks with PyTorch: Learn to build neural networks using PyTorch. It simplifies model creation and training, showcasing modern tools for scaling machine learning.
- Convolutional Neural Networks (CNNs): Discover how CNNs process image-like data through convolutional layers, pooling, and fully connected layers using PyTorch.
- Language Models for Shakespeare and Proteins: Explore sequence prediction using language models, with practical examples that apply to both text and protein sequences.
- Language Model Embeddings: Learn how to use embeddings from pre-trained models for classification and regression tasks, enhancing performance in specific applications.
- Introduction to AlphaFold: Get an overview of AlphaFold and its principles for predicting protein structures, with real-world applications.
- Graph Neural Networks (GNNs): Understand how GNNs model protein interactions and properties, providing insights into protein dynamics.
- Denoising Diffusion Probabilistic Models: Learn about diffusion models for protein structure prediction, capturing complex distributions for accurate conformations.
- Putting It All Together: Combine tools like RFdiffusion, ProteinMPNN, and AlphaFold for a comprehensive protein design workflow.
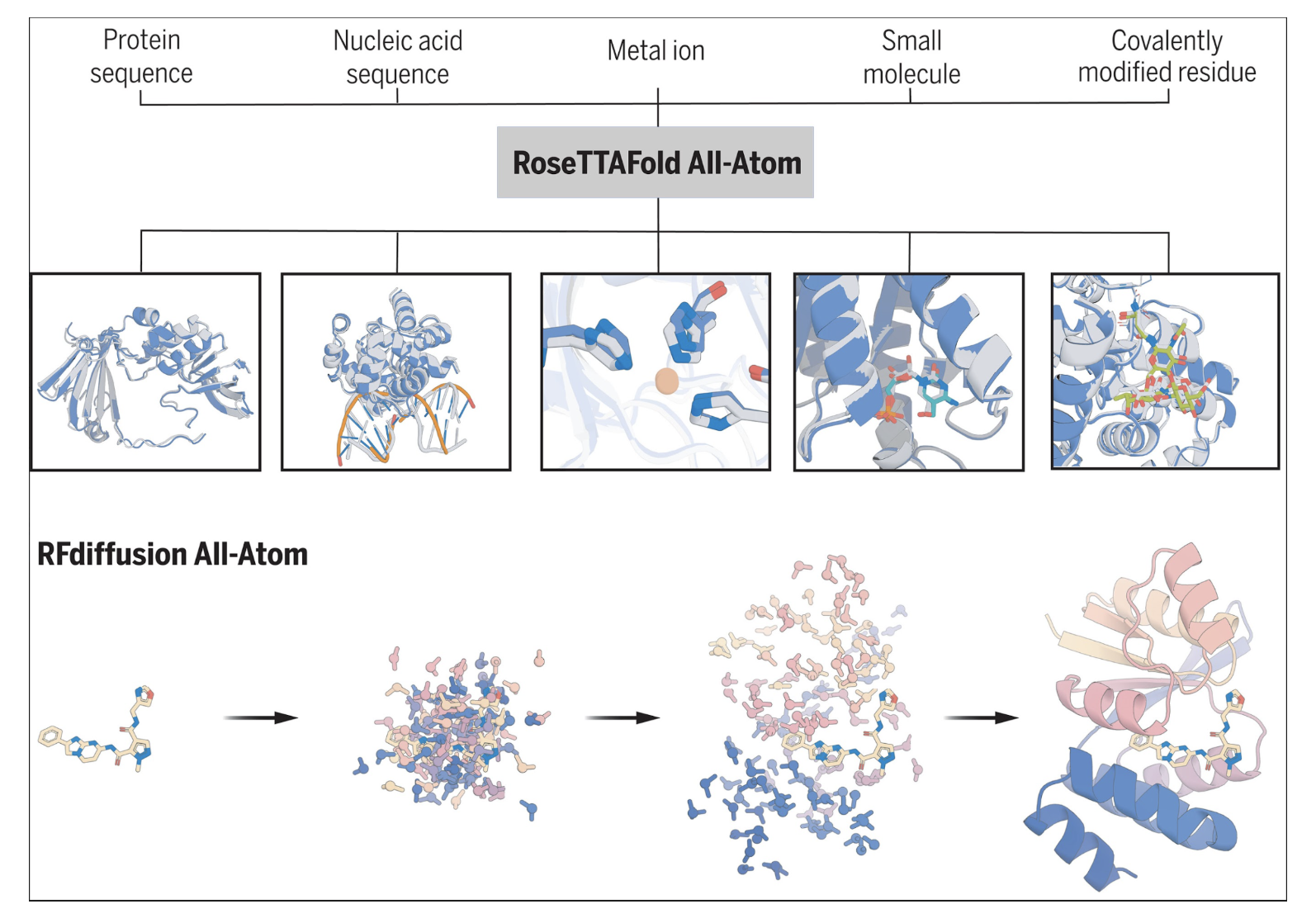
- RFDiffusion: All-Atom: Generate detailed protein structures using RFdiffusion, focusing on atomic precision.
The Value of These Resources
These notebooks provide hands-on learning opportunities for understanding and applying advanced technologies in protein structure prediction and design. They empower users to bridge the gap between machine learning concepts and practical applications in scientific research.
Conclusion
Integrating deep learning tools with protein design has great potential for advancing synthetic biology and therapeutics. Check out our GitHub Page for more resources and follow us on Twitter, Telegram, and LinkedIn for updates.
If you want to enhance your business with AI:
- Identify automation opportunities in customer interactions.
- Define KPIs for measurable impacts.
- Select AI solutions tailored to your needs.
- Implement gradually, starting with a pilot program.
For AI KPI management advice, contact us at hello@itinai.com. For continuous AI insights, follow us on Telegram and Twitter.



























